Library: Pandas¶
Installation¶
Pandas is an open-source library built on top of numpy providing high-performance, easy-to-use data structures and data analysis tools for the Python programming language.
Pandas supports the integration with many file formats or data sources out of the box (csv, excel, sql, json, parquet,…).
#!conda install pandas
#!pip install pandas
import numpy as np
import pandas as pd
labels = ['a','b','c']
my_list = [10,20,30]
arr = np.array([10,20,30])
d = {'a':10,'b':20,'c':30}
#from python list to Series
ser = pd.Series(data=my_list)
ser = pd.Series(data=my_list,index=labels)
ser = pd.Series(my_list,labels)
ser
#from Series to python list
pd.Series.to_list(ser)
#from numpy array to Series
ser = pd.Series(arr)
ser = pd.Series(arr,labels)
ser
#from Series to numpy array
pd.Series(ser).array
#from python dictionary to Series
ser = pd.Series(d)
ser
#from Series to dictionary
pd.Series.to_dict(ser)
ser = pd.Series([10,20,30],['a','b','c'])
ser
Indexing and selection¶
Pandas makes use of these index names or numbers by allowing for fast look ups of information.
ser1 = pd.Series([1,2,3,4],index = ['Enero', 'Febrero','Marzo', 'Abril'])
ser1
ser2 = pd.Series([1,2,5,4],index = ['Enero', 'Febrero','Junio', 'Abril'])
ser2
ser1['Febrero']
ser1.loc['Febrero'] #equivalent, based on label
ser1.iloc[1] #equivalent, based on position
Operations¶
Based on their associated values.
ser1 + ser2 #NaN when no matching
ser1 - ser2 #NaN when no matching
ser1 / ser2 #NaN when no matching
ser1 * ser2 #NaN when no matching
Pandas Data Frames¶
Two-dimensional, size-mutable, potentially heterogeneous tabular data.
Data structure also contains labeled axes (rows and columns). Arithmetic operations align on both row and column labels. Can be thought of as a dict-like container for Series objects.
Data Input and Output¶
Creating Data Frames¶
#passing a dictionaty with columns of different types
df = pd.DataFrame({'A': (1,2,3,4,5,6),
'B': pd.Timestamp('20130102'),
'C': pd.Series(1, index=list(range(6)), dtype='float32'),
'D': np.array([3] * 6, dtype='int32'),
'E': pd.Categorical(["test", "train", "test", "train", "test", "train"]),
'F': 'foo'})
df
Exploring the DataFrame¶
df.head(3) #first rows
df.tail(3) #last rows
df.info() #number of rows, columns, and types
df.describe() #basic description of quantitative data
df.dtypes #data type of columns
df.columns #column index names
del df['F'] #deletes column
df
df.sort_values(by='A', ascending=False) ##sorting dataframe, inplace=False by default
Selection and Indexing¶
from numpy.random import randn
np.random.seed(123)
df = pd.DataFrame(randn(5,4),index='A B C D E'.split(),columns='W X Y Z'.split())
df
#by row name
df.loc[['B']] #based on label, supports more than one
#by row position
df.iloc[[1,2]] #based on position, supports more than one
#by column name
df[['W']]
#list of column names
df[['W','Z']]
#by cell, [row,column]
df.loc['B','Y']
#by group of cell, [rows,columns]
df.loc[['A','B'],['W','Y']]
Conditional selection¶
#one condition
df[df>0] #NaN = false
#condition in a column
df[df['W']>0]
#condition in a column, select other columns
df[df['W']>0][['Y','X']]
#two conditions in different columns
df[(df['W']>0) & (df['Y'] > 1)] #being & = AND
#one condition or the other in different columns
df[(df['W']>0) | (df['Y'] > 1)] #being | = OR
Modify index¶
# Reset to default 0,1...n index
df.reset_index()
df
#new index
nind = 'blue green purple red yellow'.split()
df['Colours'] = nind
df.set_index('Colours')
#df.set_index('Colours',inplace=True) #to make it permanent
df
Creating new column¶
df['newColumn'] = df['W'] + df['Y']
df
Droping column¶
df.drop('newColumn',axis=1, inplace=True) #inplace=True makes it permanent
df.drop('Colours',axis=1, inplace=True) #inplace=True makes it permanent
df
df.drop('E',axis=0) #inplace=True makes it permanent
df
Null values¶
df = pd.DataFrame({'A':[1,2,np.nan],
'B':[5,np.nan,np.nan],
'C':[1,2,3]})
df
df.isnull() #finding nulls
df.isna() #finding na
df.dropna() #drops very row with NaN
df.dropna(axis=1) #drops every column with NaN
df.dropna(thresh=2) #drops every row with specific number of NaN
df.fillna(value='FILL VALUE') #replaces NaN with a defined value
df['A'].fillna(value=df['A'].mean()) #replaces NaN with the mean of the column
Multi-Index Hierarchy¶
#Creates a data frame with multi-index
outside = ['bar', 'bar', 'baz', 'baz', 'foo', 'foo', 'qux', 'qux'] #index generated
inside = ['one', 'two', 'one', 'two', 'one', 'two', 'one', 'two'] #index generated
hier_index = list(zip(outside, inside)) #indexs are zip together
hier_index = pd.MultiIndex.from_tuples(hier_index, names=['first', 'second']) #hierarchy of index applied
df = pd.DataFrame(np.random.randn(8,3), index=hier_index, columns= ['A', 'B', 'C']) #random data generated
df
#change index names
df.index.names
df.index.names = ['first','second']
df
df.loc['foo'] #outer index selected
df.xs('foo') #equivalent
df.loc['foo'].loc['one'] #outer and inner index selected by name
df.xs(['foo','one']) #equivalent
df.xs('two',level='second') #cross-section function allows indexing directly inner index
Groupby¶
The groupby method allows to group rows of data together and call aggregate functions.
# Create dataframe
data = {'Company':['GOOG','GOOG','MSFT','MSFT','FB','FB'],
'Person':['Sam','Charlie','Amy','Vanessa','Carl','Sarah'],
'Sales':[200,120,340,124,243,350]}
df = pd.DataFrame(data)
df
df.groupby("Company") #group using criteria
df.groupby("Company").mean() #select way of aggregation(ex. mean)
df.groupby("Company").sum() #sum of the group
df.groupby("Company").max() #maximum of the group
df.groupby("Company").std() #standard deviation of the group
df.groupby("Company").count() #count cases of the group
df.groupby("Company").describe().transpose() #describe + transpose
Merging, Joining, and Concatenating¶
df1 = pd.DataFrame({'A': ['A0', 'A1', 'A2', 'A3'],
'B': ['B0', 'B1', 'B2', 'B3'],
'C': ['C0', 'C1', 'C2', 'C3'],
'D': ['D0', 'D1', 'D2', 'D3']},
index=[0, 1, 2, 3])
df1
df2 = pd.DataFrame({'A': ['A4', 'A5', 'A6', 'A7'],
'B': ['B4', 'B5', 'B6', 'B7'],
'C': ['C4', 'C5', 'C6', 'C7'],
'D': ['D4', 'D5', 'D6', 'D7']},
index=[4, 5, 6, 7])
df2
Concatenate¶
Dimensions should match along the axis you are concatenating on, gets all the rows together.
pd.concat([df1,df2], axis=0, join='outer', ignore_index=False, keys=None,
levels=None, names=None, verify_integrity=False, copy=True)
result = df1.append(df2) #equivalent
result
Merge¶
Allows you to merge DataFrames together using a similar logic as merging SQL Tables together.
#creates left table
left = pd.DataFrame({'key': ['K0', 'K1', 'K2', 'K3'],
'A': ['A0', 'A1', 'A2', 'A3'],
'B': ['B0', 'B1', 'B2', 'B3']})
left
#creates right table
right = pd.DataFrame({'key': ['K0', 'K1', 'K2', 'K3'],
'C': ['C0', 'C1', 'C2', 'C3'],
'D': ['D0', 'D1', 'D2', 'D3']})
right
#performs INNER merge (only coincidences in key between dataframes are kept)
pd.merge(left, right, how='inner', on='key', left_on=None, right_on=None,
left_index=False, right_index=False, sort=True,
suffixes=('_x', '_y'), copy=True, indicator=False,
validate=None)
#creates left table
left = pd.DataFrame({'key1': ['K0', 'K0', 'K1', 'K2'],
'key2': ['K0', 'K1', 'K0', 'K1'],
'A': ['A0', 'A1', 'A2', 'A3'],
'B': ['B0', 'B1', 'B2', 'B3']})
#creates left table
right = pd.DataFrame({'key1': ['K0', 'K1', 'K1', 'K2'],
'key2': ['K0', 'K0', 'K0', 'K0'],
'C': ['C0', 'C1', 'C2', 'C3'],
'D': ['D0', 'D1', 'D2', 'D3']})
#performs inner merge (only coincidences between dataframes in both keys are kept)
pd.merge(left, right, on=['key1', 'key2'])
#performs OUTER merge (all cases are kept)
pd.merge(left, right, how='outer', on=['key1', 'key2'])
#performs RIGHT merge (all cases of right dataframe are kept)
pd.merge(left, right, how='right', on=['key1', 'key2'])
#performs LEFT merge (all cases of left dataframe are kept)
pd.merge(left, right, how='left', on=['key1', 'key2'])
Join¶
Combining the columns of two potentially differently-indexed DataFrames into a single result DataFrame.
left = pd.DataFrame({'A': ['A0', 'A1', 'A2'],
'B': ['B0', 'B1', 'B2']},
index=['K0', 'K1', 'K2'])
right = pd.DataFrame({'C': ['C0', 'C2', 'C3'],
'D': ['D0', 'D2', 'D3']},
index=['K0', 'K2', 'K3'])
left.join(right) #inner join
left.join(right, how='outer') #outer join
Operations¶
#creates data frame
df = pd.DataFrame({'col1':[1,2,3,4],'col2':[444,555,666,444],'col3':['abc','def','ghi','xyz']})
df
#Select from DataFrame using criteria from multiple columns
newdf = df[(df['col1']>2) & (df['col2']==444)]
newdf
#list of unique values
df['col2'].unique()
#number of unique values
df['col2'].nunique()
#amount of each value
df['col2'].value_counts()
#Applying created functions
def times2(x):
return x*2
df['col1'].apply(times2)
#Applying standard functions
df['col3'].apply(len) #lenght of the values
df['col1'].sum() #sum of the values
Pivot table¶
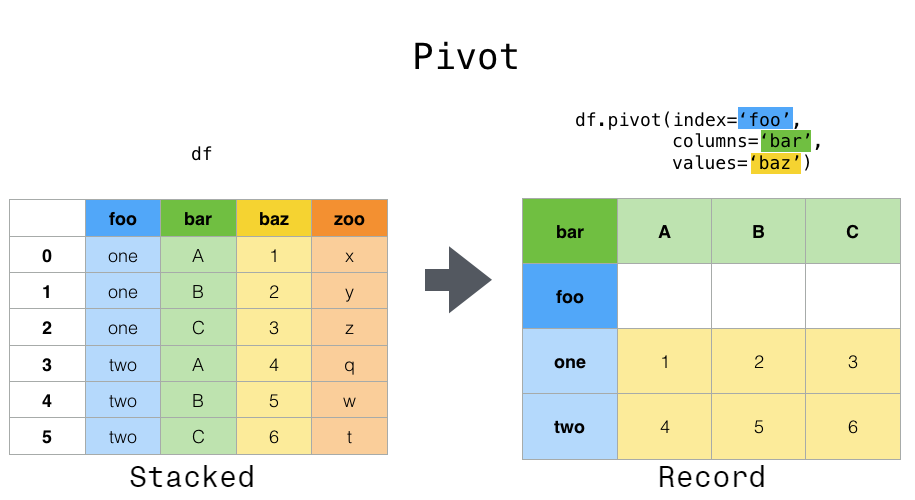
#creates data frame from dictionary
data = {'A':['foo','foo','foo','bar','bar','bar'],
'B':['one','one','two','two','one','one'],
'C':['x','y','x','y','x','y'],
'D':[1,3,2,5,4,1]}
df = pd.DataFrame(data)
df
#creates pivot table
df.pivot_table(values='D',index=['A', 'B'],columns=['C'])
Reshaping by stacking¶
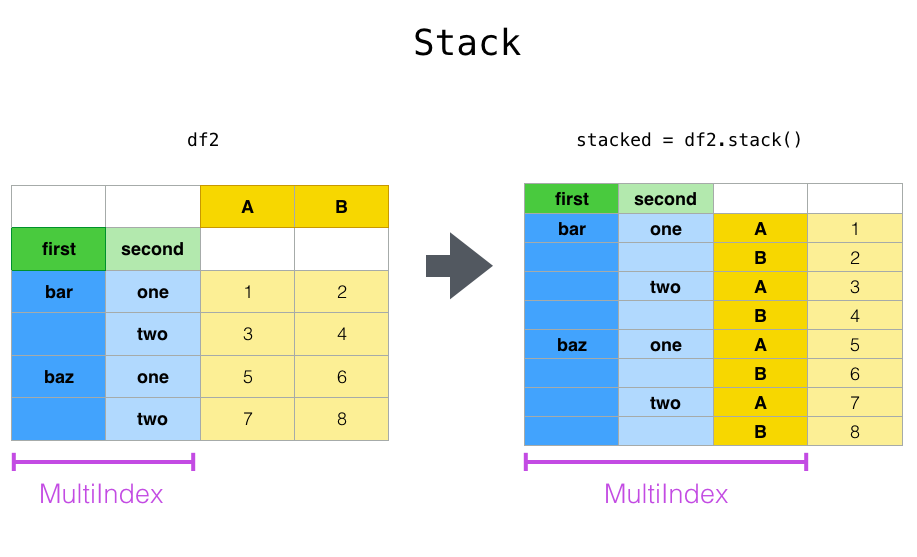
df
df2 = df[:4]
df2
#Stacking
stacked = df2.stack()
dfstacked = pd.DataFrame(stacked, columns =['cases'])
dfstacked
#Unstacking
dfstacked.unstack()
Reshaping by Melt¶
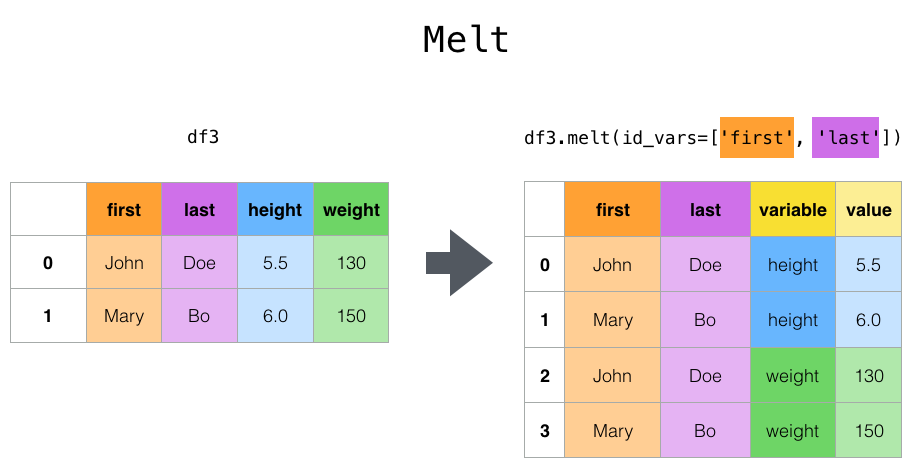
cheese = pd.DataFrame({'first': ['John', 'Mary'],
'last': ['Doe', 'Bo'],
'height': [5.5, 6.0],
'weight': [130, 150]})
cheese
cheese.melt(id_vars=['first', 'last'], var_name='quantity')
Data visualization¶
import matplotlib as plt
import seaborn as sns
# loading databases
tips = sns.load_dataset('tips')
tips.head()
%matplotlib inline
Styles¶
#set matplotlib style
plt.style.use('default')
tips['total_bill'].hist()
plt.style.use('ggplot')
tips['total_bill'].hist()
plt.style.use('bmh')
tips['total_bill'].hist()
plt.style.use('fivethirtyeight')
tips['total_bill'].hist()
plt.style.use('classic')
tips['total_bill'].hist()
plt.style.use('grayscale')
tips['total_bill'].hist()
plt.style.use('seaborn')
tips['total_bill'].hist()
Graphic types¶
df1 = tips[0:20][['total_bill', 'tip']]
df1.head()
df2 = tips[['total_bill', 'tip', 'day']]
df2.head()
df3 = tips[0:20][['total_bill', 'tip', 'size']]
df3.head()
df1.plot.area() #can only use quantitative information, qualitative make error
df1.plot.barh(stacked=True) #can only work with quantitative data, skips qualitative
df1.plot.density() #distribution of quantitative data, skips qualitative
df2['total_bill'].plot.hist(bins = 10) #distribution of quantitative data, skips qualitative
df2.plot.line(figsize=(12,3),lw=1) #works with quantitative data, skips qualitative
df3.plot.scatter(x = 'total_bill', y = 'tip', c = 'size', s=df3['size']*100, cmap = 'rainbow') #can only use quantitative information, qualitative make error
df1.plot.bar(stacked = True) #can only work with quantitative data, skips qualitative
df1.plot.box() #categories are the columns, can be used "by = " for groupby
df1.plot.hexbin('total_bill', 'tip',gridsize=25,cmap='Blues') #distribution of quantitative data, skips qualitative
df1.plot.kde()